COMPUTER-AIDED MOLECULAR DESIGN
CCG is a leading developer and provider of Molecular Modeling, Simulations and Machine Learning software to Pharmaceutical and Biotechnology companies as well as Academic institutions throughout the world. CCG continuously develops new technologies with its team of mathematicians, scientists and software engineers and through scientific collaborations with customers.
MOE
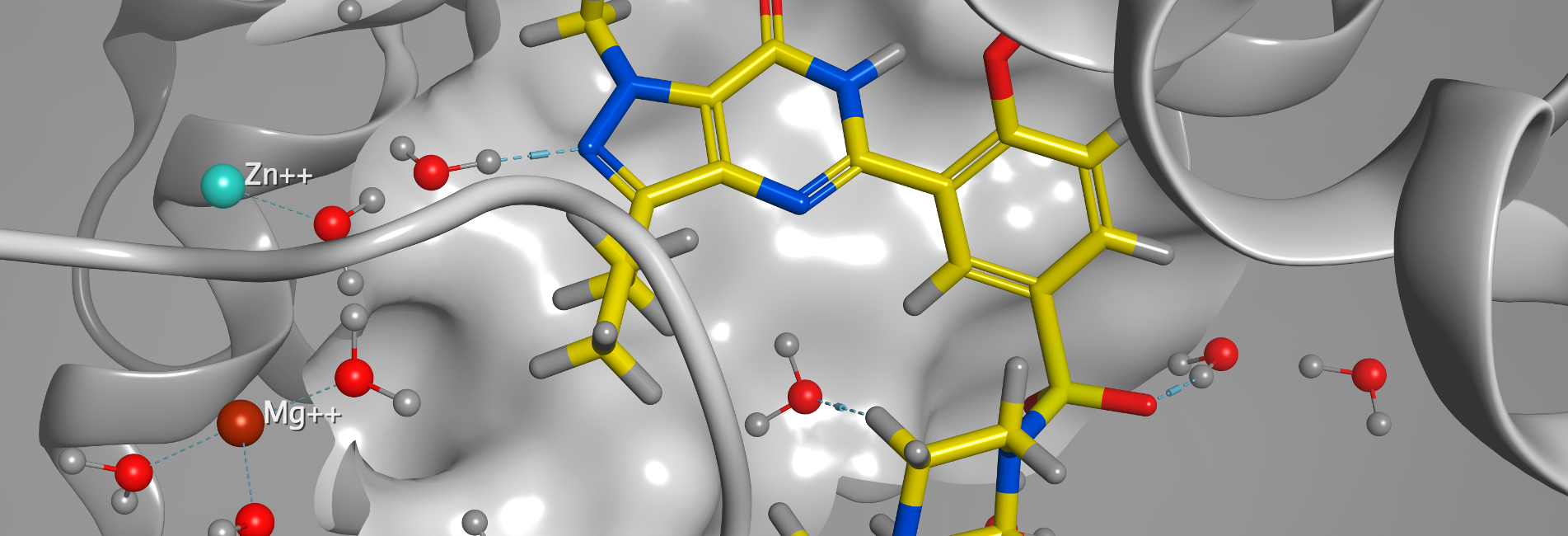
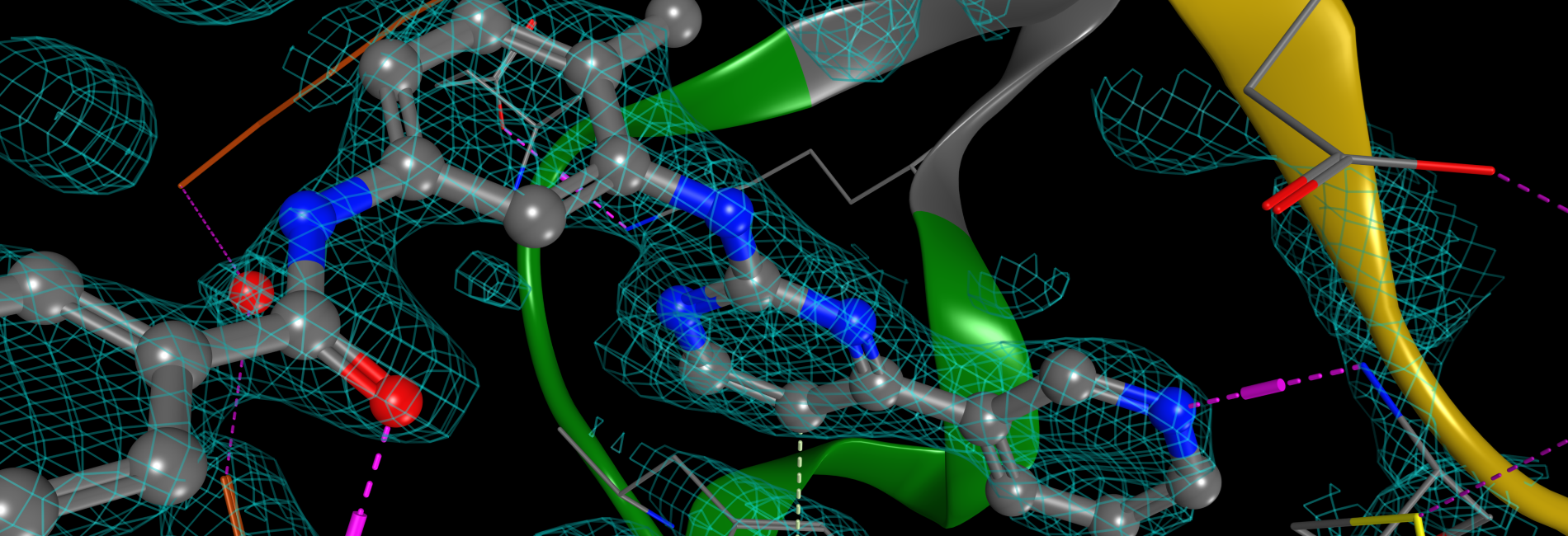
Structure-Based Drug Design
Ligand- and Fragment-Based Drug Design
Pharmacophore Screening
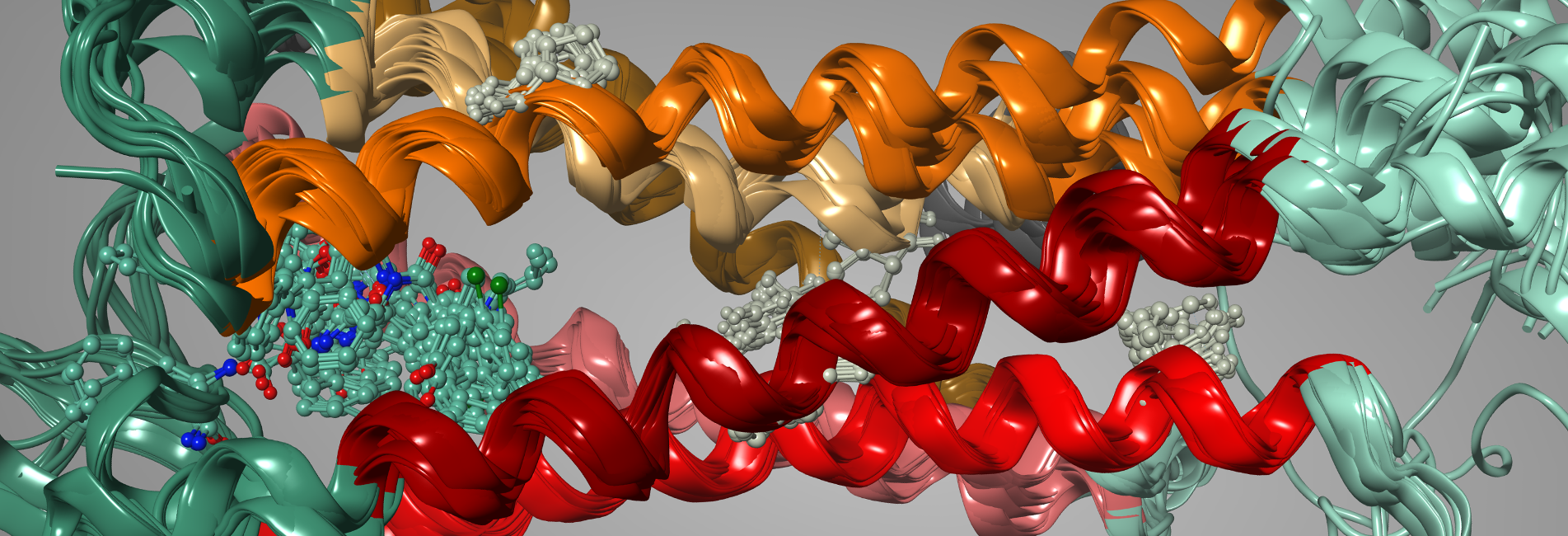
Protein, Antibody and Peptide Modeling
Molecular Modeling and Simulations
Cheminformatics and QSAR
Methods Development and Deployment
Integrated Discovery Platform
MOEsaic
Structure-Activity and Property Relationships
Visualize SAR Data and Trends
Perform Substructure and Similarity Searches
Profile and Analyze R-Groups
Analyze Matched Molecular Pairs (MMPs)
Design Novel Virtual Ligands
Document Analysis Results and Collect Notes
Web-Based SAR Explorer
PSILO
Macromolecular Repository
Data Visualization and Analysis
Browser Interface for Search and Retrieval
3D Ligand: Receptor and Pocket Similarity
Protein Structure Alignment
Data Standardization and Annotation
Standard IT Infrastructure and Integration